Microbial Bioinformatics
How does a bacterial cell work? Wich bacterial species is responsible for a certain function in a catalytically active biofilm? How do persistent infections and antibiotic resistances develop? There is a variety of ways to deal with these questions and to answer them, it is usually necessary to link different disciplines.
The term ‘bioinformatics’ generally is meant to describe an interdisciplinary methodology that analyses biological questions by applying methods and algorithms originating from mathematics and informatics. In contrast to ‘classical’ wet lab methods, a more holistic approach is usually applied that treats experimental subjects as a whole (especially in ‘systems biology’). Besides the genome – the whole of all genes within an organism – the suffix ‘omics’ has now been used for various ‘wholes’, e.g., the transcriptome, the proteome, the metabolome or the interactome.
Modern sequencing technologies develop at a high pace, enabling the analysis of bacterial genomes and their expression with a depth and accuracy, which seemed unbelievable just a decade ago. At the same time, the cost per sequenced base pair has constantly diminished and today, many research institutions already have access to the latest sequencing technology, themselves. While the sequencing of a bacterial genome is therefore easily affordable and quickly achieved, the analysis of the resulting ‘omics’-data remains the actual challenge.
The group ‘microbial genomics and bioinformatics concentrates on the meaningful interpretation of the large amounts of newly available data within the background of existing knowledge. Additionally, we focus on the development of new hypotheses and methods that emerge from the new technologies and insights.
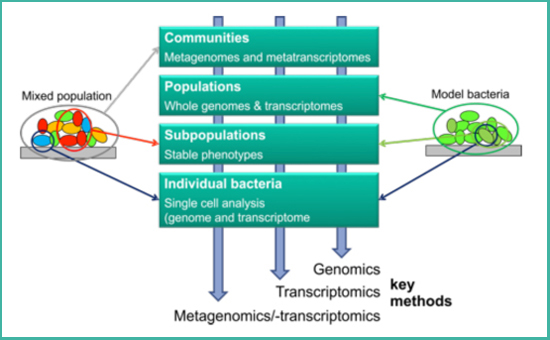
(Fig. 1) Genomes and transcriptomes can be examined on different levels: bacterial populations consist of different subpopulations and these in turn of individual bacterial cells; different species may be part of microbial communities.
Scientific focus:
- Evaluation of Next Generation Sequencing (NGS) data and other ‘omics’ data
- Analysis of bacterial genomes and transcriptomes
- Characterization of bacterial communities by metagenomics and metatranscriptomics